Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Comparison of Three Delivery Modes for CXCR4 Excision via CRISPR/Cas9 System
*Corresponding author: Xiangwen Peng, Hunan Provincial Key Laboratory of Regional Hereditary Birth Defects Prevention and Control, Changsha Hospital for Maternal & Child Health Care, Hunan Normal University, No. 416 East Chengnan Road, Yuhua District, Changsha 410001, PR China and Lei Shi, Institute of Food Safety and Nutrition, Jinan University, Guangzhou 510632, PR China.
Received: October 04, 2024; Published: October 14, 2024
DOI: 10.34297/AJBSR.2024.24.003198
Abstract
Background: Host innate immune response has been proved to be affected by gene therapy. Host immune factors are essential for antagonizing human immunodeficiency virus type 1 (HIV-1) infection. Endogenous CXCR4 and CCR5, two human chemokine receptors, were evident to assist HIV for the invasion into human cells by membrane fusion at the initial viral infection stage.
Methods: Previously, studies have demonstrated that mutations of co-receptors (CCR5 and/or CXCR4) can endow host cells with resistance for the HIV-1 infection. It is common that programmable RNA-guided CRISPR/Cas9 complex is designed to target and mutate the cellular genomic sequences, providing the ability to resist HIV. Thus, this novel technology has the potential therapeutic roles for HIV infection through gene editing of candidate genes.
Results: Here in this study, we demonstrated that, by performing CRISPR/Cas9 system equiped with CXCR4-targeted sgRNAs, the expression of co-receptor CXCR4 can be ablated by Cas9 endonuclease via three different delivery strategies, conferring resistance to HIV-1 infection. Our results suggested that electroporation of CRISPR/Cas9 ribonucleoproteins (ECRNP), the most efficient method for silencing CXCR4 gene, induces indels with frequencies of up to 82%, following by 68% of lentiviral vectors encoding CRISPR/Cas9 (Lenti-CRISPR/Cas9) and 38% of lipofectaminedelivered CRISPR/Cas9 plasmids (LDCP).
Conclusions: The CRISPR/Cas9-mediated silence of a certain host gene (e.g., CXCR4) offer an alternative path to modulate host immunity for combating against HIV-1 infection. Importantly, the effective delivery system is crucial for knocking out/down CXCR4 gene and mitigating off-target events.
Keywords: CCR5; CRISPR/Cas9; CXCR4; Delivery vector; HIV-1; Off-target
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 (CRISPR-associated nuclease 9) have sparked a new biological revolution worldwide since its development in 2013 [1,2]. CRISPR/Cas9, a defence mechanism firstly found in prokaryotes, is generally a binary complex architecture made up of a sequence-specific guide RNA (sgRNA) and a non-specific Cas9 endonuclease. Recently, this two-component system has been developed into a cost-effective, easy-operating, and highly specific targeted genome editing platform, which provides an innovative potential medical method for treating viral infectious diseases, including chronic HIV infection. During the treatment of HIV/AIDS, precise cell-targeted delivery of CRISPR/Cas9 pharmaceutical reagents is one of determinants for effective HIV-1 elimination. In addition, key candidate host genes, CCR5 (C-C chemokine receptor type 5) and CXCR4 (C-X-C chemokine receptor type 4), serves as two effective gene therapeutic targets for engineering host immune resistance against HIV-1 infection [3]. This is because that molecularly, in order to binding to CD4, one of the host cell co-receptor (CXCR4 or CCR5) is required for HIV to fuse the cellular membrane through activation of gp41 (gp41, an HIV envelope protein) and enter into the cells [4]. This causes the release of the viral genetic materials into the extracellular matrix for the next round of infection. Normally, CCR5 acts as a HIV co-receptor for assisting virus entering into cells at early stage; while CXCR4 receptor in CXCR4-tropic HIV strains mainly emerges at late stages of infection, mediating cell entry and contributing to disease progression [5,6]. Homozygous CCR5-Δ32 gene deletion in people can beneficially enhance their immune system and defense the infection of HIV [7]. Additionally, as a most important receptor of stromal cell-derived factor-1 (SDF-1), CXCR4 is expressed in multiple cell types (e.g., cardiac cells, brain cells, lung cells, and kidney cells), involving in many pathophysiological processes such as assimilate, metabolism and immunity [8]. Therefore, modifying host-dependent factors to adjust host immunity can be a more effective treatment strategy for HIV-1 elimination than targeting the viral essential elements on virus, owing to the potential dormant state of HIV-1 within the host genome [9]. More importantly, directly targeting virus has many deficiencies, especially during the step of eliminating latent proviral DNA of HIV-1 in vitro, the viral escape [10,11] and even the mutation and diversity of HIV-1 genetic information [12-14] can be facilitated.
Based on the selectivity of HIV for co-receptor in host cells, HIV-1 strains can be classified into CCR5 (R5), CXCR4 (X4), or dual-tropic. Previous preclinical studies have elucidated that, by conducting CRISPR/Cas9 system, genetic modifications to mutate HIV co-receptors CXCR4 and/or CCR5 in human CD+ T cells in vitro can resist HIV and prevent infection [15-18]. Nevertheless, CRISPR/ Cas9-based applications in the treatment of HIV/AIDS disease still faces several challenges. Firstly, HIV-1 is resistant to most inhibitors that target its constituent parts, including CRISPR/Cas9 treatment approaches [19]. In details, the tricky HIV can circumvent host adaptive immunity and easily generate viral mutations to resist CRISPR/Cas9-mediated elimination in vivo. Secondly, the CRISPR/ Cas9 systems that specifically targeted CXCR4 and/or CCR5 should be modified to upgrade CRISPR/Cas9 systems, maybe through designs of less off-targeted sgRNAs. Thirdly, the delivery vehicles for CRISPR/Cas9 complexes remain to be further developed, no matter what type of vectors, viral or non-viral.
Most recently, Turk, et al. reported that a sterilizing cure of HIV- 1 infection has been discovered in two patients with HIV-1 who accept allogeneic hematopoietic stem cell transplantations from donors that naturally have homozygous CCR5Δ32 gene [20]. This is an extremely rare event for HIV sterilizing cure but do provide a possible outcome of HIV-1 infection. However, low efficiency of vector-depended genome editing via homology-directed repair (HDR) or non-homologous end joining (NHEJ) repair leads to a serious obstacle to the use of CRISPR/Cas technology in clinical practice [21]. In recent years, researchers have revealed that HIV co-receptor factors (mainly CCR5 and CXCR4) can be targeted successfully using CRISPR/Cas9 in human cell strains (e.g., primary CD4+ T cells) in vitro for fighting against HIV-1 infection [22-24]. The delivery of CRISPR/Cas9 system via optimized ribonucleoprotein (RNP) can be a novel strategy for the therapeutic treatment for HIV-1 infection. However, according to data analysis, the detail mechanisms of anti-HIV bioactivity in vivo is still elusive, and drug development to cure AIDS is still struggling.
In the present study, we employed the CRISPR/Cas9 system to target and incapacitate cellular co-receptor CXCR4 gene for conferring anti-HIV effects in HIV-1 susceptible human primary CD4+ T cells through different Cas9 delivery modes, including lipofectamine-delivered CRISPR/Cas9 plasmids (LDCP), lentiviral plasmids encoding CRISPR/Cas9 (Lenti-CRISPR/Cas9) and direct electroporation of CRISPR/Cas9 ribonucleoproteins (ECRNP). The gene mutation efficacy was detected and assessed by referring the expression of T7 endonuclease I (T7E1) among different groups. Our data demonstrated that design and optimization of package and delivery modes for CRISPR/Cas9 system have significant implications for in vitro gene therapy of HIV-1 infection.
Materials and Methods
Construction of Plasmids
CRISPR/Cas9 plasmids were constructed by restriction endonucleases and annealed oligonucleotides which containing a 20-base leading sequence fragment. The annealed oligonucleotides were ligated into the pX330 plasmid that contains a U6 promoter-driven chimeric guide strand and a CBh promoter-driven Cas9 expression cassette via the sticky ends of oligonucleotides. Thus, Streptococcus pyogenes CRISPR/Cas9 (SpCRISPR/Cas9) system was co-expressed from the 8484-bp Cas9 gene expression plasmid, pX330 (Addgene plasmid #42230, pX330-U6-Chimeric_BB-CBh-hSpCas9).
Lentiviral packaging plasmid (psPax2; Addgene #12260) was obtained from the Addgene repository (Water-town, MA, USA). Lentivirus expressing Cas9 and CXCR4 sgRNA (Lenti‑CRISPR/ Cas9) was constructed using the form of “all-in-one” viral vector as previously. The sgRNA carrying crRNA-loop-tracrRNA for targeting CXCR4 were designed and synthesized as previously described [25,26]. To generate constructs to target CXCR4, the CXCR4-targeted sgRNAs containing crRNA-loop-tracrRNA was inserted into the lenti-CRISPR/Cas9 vector. Among them, two available CXCR4-directed sgRNAs were screened from twelve designed candidates (Figure 1) based on the former study [18]. The sgRNA design was used online design software (https://zlab.bio/guide-design-resources). Constructed plasmids were confirmed by Sanger sequencing.
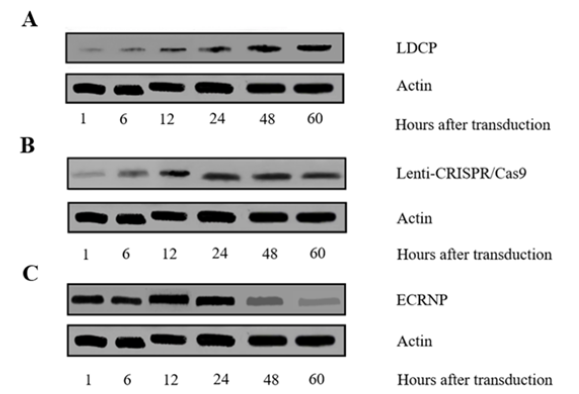
Figure 1: The protein level of Cas9 in HEK-293T cells at indicated times (1 h, 6 h, 12 h, 24 h, 48 h, and 60 h) after transduction with LDCP, Lenti-CRISPR/Cas9 and ECRNP, respectively, and were determined by western blotting analysis. (A) Cas9 protein was prominent at 48 h in LDCP delivery pattern. (B) Cas9 protein was prominent at 24 h after transduction with Lenti-CRISPR/Cas9. (C) Cas9 protein transported with ECRNP mode was expressed continually in a high-level within 24 h, but from 24 h to 60 h, its expressions had declined gradually.
Cell Culture and Transduction
The sealed packaging HEK-293T cells were purchased from Invitrogen Life Technologies and maintained in Dulbecco’s modified Eagle’s medium (DMEM) medium (high glucose DMEM supplemented with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin, 100 mg/ml streptomycin) plus puromycin (1 μg/ml) in a 10-cm cell culture dish.
TZM-bl cells (recombinant HeLa cells expressing high levels of CD4 receptor, CXCR4 and CCR5 co-receptors) were obtained and cultured as described previously [27, 28]. HeLa-cell derived HIV-1 reporter TZM-bl cells were maintained in Dulbecco’s Medium supplemented with 10% FBS, 100 U penicillin ml−1 and 100 mg streptomycin ml−1 at 37°C and 5% CO2. TZM-bl cells (3~5×105) were co-transfected with 2μg of CRISPR/Cas9-gRNA plasmid and 2μg of targeting vector using lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA). After 3 days transfection, the cells were selected by puromycin (1μg/ml) for 2 weeks with daily medium change. Then, single drug-resistant clones were picked and propagated in 12-well plates for further analysis.
Human peripheral blood mononuclear cells were obtained from Zen-Bio (Research Triangle Park, Durham, NC, USA). CD4 T cells were isolated from peripheral blood mononuclear cells using CD4 T cell enrichment kit (Stem Cell Technologies, Vancouver BC, Canada). Human CD4+ T cells were stimulated with CD3/CD28 (Thermo-Fisher Scientific, Waltham, MA, USA) before transduction with lentivirus and were cultured at 37°C in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100U/ml of penicillin-streptomycin with recombinant IL-2 (20U/ml).
CRISPR/Cas9 Ribonucleoproteins (RNPs) Assembly and Electroporation
Cas9 RNPs were prepared swiftly prior to using in experiments by incubating 20 μM Cas9 enzyme with 20 μM sgRNA at ratio of 1:1 in 20 μM HEPES (hydroxyethyl piperazine ethanesulfonic acid) buffer (pH 7.5), 150 mM KCl, 1 mM MgCl2, 10% (vol/vol) glycerol, and 1 mM Tris (β-chloroethyl) phosphate (TCEP) at 37°C for 10 min to a final concentration of 10 μM. Primary T cells were electroporated with a Neon® Transfection system 100 μL Kit and Thermo Invitrogen Neon device.
T7E1 Assay
Genomic DNA was isolated from target cells using the QIAamp Micro DNA kit (Qiagen, Waltham, MA) following in vitro transfection with CRISPR/Cas9 systems. Genome editing events caused by CRISPR/Cas9 system packaged in three different modes were quantified by applying the EnGen® Mutation detection kit (New England Biolabs) according to the manufacturer’s instructions.
Western Blot Analysis
The cell lysates were obtained from control groups and experimental groups (Cas9-treated HEK-293T cells using three different delivery vectors) in the presence of protease inhibitors. The protein samples were resolved by SDS-PAGE (4~12%) and electroblotted onto a nitrocellulose membrane. The membrane was blocked with skim milk (Block Ace®). The membrane was probed with anti-caspase 3 antibody (×1/750) and then POD-tagged anti-mouse IgG (gamma) antibody (×1/1,500). The membrane was soaked in ECL (electrochemiluminescence) solution and then wrapped. After putting the wrapped membrane in a film cassette, an X-ray film (Hyper film®, GE Healthcare) was developed in a dark place.
Flow Cytometry Analysis
Flow cytometry was employed to determine the relative knockout efficacy of CXCR4 gene. Briefly, 2.5×105 modified or control cells were collected and washed three times in 1×PBS, and then the cells were stained with PE-conjugated anti-CXCR4 for 15 min at 4°C. The cells were then washed for three times after incubation. CXCR4 knock-out was determined by analyzing the CXCR4 expression levels via flow cytometry (BD FACS Aria III) and by PCR at the genetic level followed by T7E1 digestion. The data were analyzed with FlowJo v10 software (Treestar).
Statistical Analysis
All data are presented as the mean ± SD (standard deviation). Statistical analysis was conducted by using SPSS 19.0 in Windows 11. Comparisons between two groups were performed by statistics method Paired Student’s t-tests. Comparisons among groups were analyzed by one-way ANOVA test. Values of P<0.05 were considered to be statistically significant. All experiments were performed repeat no less than three times.
Results
The Efficient Cas9 Protein Delivery by ECRNP
In comparison to the prolonged plasmid expression of Cas9, protein transduction offers a notable reduction in off-target effects. Our study investigated the Cas9 protein expression in HEK-293T cells over different time points post-transduction through western blot analysis (Figure 1). The efficiency of Cas9 protein delivery (ECRNP) surpassed that of Cas9 plasmid expression methods (LDCP and Lenti-CRISPR/Cas9) for gene editing in HEK-293T cells. Notably, LDCP and Lenti-CRISPR/Cas9 exhibited similar trends (Figure 1A, B), with gradually increasing protein levels over time. ECRNP demonstrated a rapid emergence, reaching its peak at 24 hours post-transduction, followed by a decline in expression from 48 hours onwards (Figure 1C).
The Knockdown of CXCR4 in TZM-bl Cells by CRISPR/Cas9 Delivery System
In theory, genomic sequences following the pattern 5′-GN19NGG-3′ (or 5′-CCNN19C-3′) are potential targets for the SpCRISPR/Cas9 system. To genetically disrupt the HIV co-receptor gene CXCR4, we devised three distinct delivery methods (LDCP, Lenti-CRISPR/Cas9, and ECRNP) for the SpCRISPR/Cas9 system in TZM-bl cells (Figure 2A). Utilizing the online web server (http:// crispr.mit.edu), we designed twelve single-stranded gRNA sequences targeting the second exon of the CXCR4 gene (NM_003467) (Figure 2B). Then, the TZM-bl cell line of epithelial origin and Lenti-CRISPR/Cas9 delivery mode were chosen to assess the efficacy of CRISPR/Cas9 system among the 12 sgRNA alternatives for inducing CXCR4 co-receptor ablation. Genomic DNA extracted from the cells was PCR amplified, and T7E1 assay, coupled with a 1.5% agarose gel, was employed to determine CXCR4 disruption (Figure 2C). In the process, puromycin served as the selection marker for screening stable expression cell lines. To eliminate unedited target cells and enhance transfection efficiency, TZM-bl cells were incubated with 1μg/ml puromycin for 24 hours. T7E1 assay results indicated the effectiveness of five out of twelve sgRNAs in Lenti-CRISPR/ Cas9-mediated CXCR4 disruptions in TZM-bl cells. Our results suggested that sgRNA #1 and sgRNA #2 had higher efficiency in CXCR4 knockdown compared to the other three (Figure 2C, 2D). The specific targeted sequences of the custom-designed sgRNAs used in the experiment are listed in Figure 2E.
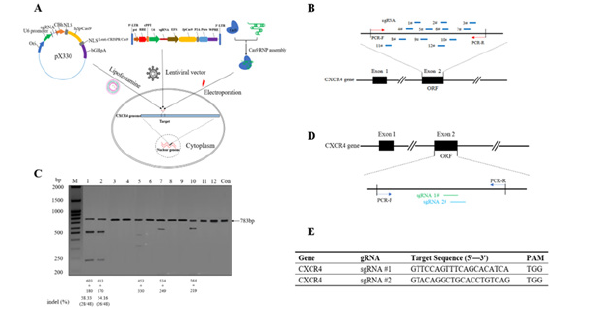
Figure 2: Schematic diagram of CRISPR/Cas9 technology mediated mutation of CXCR4 locus. (A) Schematic diagram of three delivery modes for CRISPR/Cas9 system (LDCP, Lenti-CRISPR/Cas9 and ECRNP) to mediate genome editing of CXCR4 locus, respectively. (B) Schematic of the gRNAs for targeting CXCR4 gene locus. CXCR4-tageted gRNAs are shown by blue box. The specific PCR primer pairs are shown in red arrows. Forward and reverse primers for PCR are shown in red arrows to denote the amplify the region. (C) T7EN1 assay for cleavage activities of twelve CXCR4-targeted sgRNAs coupling with Lenti-CRISPR/Cas9 respectively in TZM‑bl cells. The TZM‑bl cells were infected with specified CXCR4-sgRNA lentivirus plasmid at an M.O.I. of 40. The migrating bands below 783bp in lanes indicate the disrupted CXCR4 alleles. (D) Schematic of two screened sgRNAs for targeting CXCR4 coding region in human genomic DNA sequences. (E) Two sgRNAs for CRISPR/Cas9 system and their targeting sequences.
Fig. 2. Schematic diagram of CRISPR/Cas9 technology mediated mutation of CXCR4 locus. (A) Schematic diagram of three delivery modes for CRISPR/Cas9 system (LDCP, Lenti-CRISPR/Cas9 and ECRNP) to mediate genome editing of CXCR4 locus, respectively. (B) Schematic of the gRNAs for targeting CXCR4 gene locus. CXCR4-tageted gRNAs are shown by blue box. The specific PCR primer pairs are shown in red arrows. Forward and reverse primers for PCR are shown in red arrows to denote the amplify the region. (C) T7EN1 assay for cleavage activities of twelve CXCR4-targeted sgRNAs coupling with Lenti-CRISPR/Cas9 respectively in TZM‑bl cells. The TZM‑bl cells were infected with specified CXCR4-sgRNA lentivirus plasmid at an M.O.I. of 40. The migrating bands below 783bp in lanes indicate the disrupted CXCR4 alleles. (D) Schematic of two screened sgRNAs for targeting CXCR4 coding region in human genomic DNA sequences. (E) Two sgRNAs for CRISPR/Cas9 system and their targeting sequences.
CRISPR/Cas9-Mediated Editing of CXCR4 Gene in Human CD4+ T Cells
In this study, we examined the impact of employing three distinct vectors to deliver the CRISPR/Cas9 system for CXCR4 deletion in human CD4+ T cells. Human CD4+ T cells were transfected with control, #1, or #2 CXCR4-gRNA/Cas9, and T7EN1 assays were conducted three days post-transfection. Our observations revealed that both #1 and #2 gRNA, in conjunction with Cas9 endonucleases, induced indel mutations in the CXCR4 gene within primary CD4+ T cells (Figure 3A). The flow cytometry (FC) and DNA sequences were employed to analyse the effects of different CRISPR/Cas9 systems transported by different modes (Figure 4B, C).
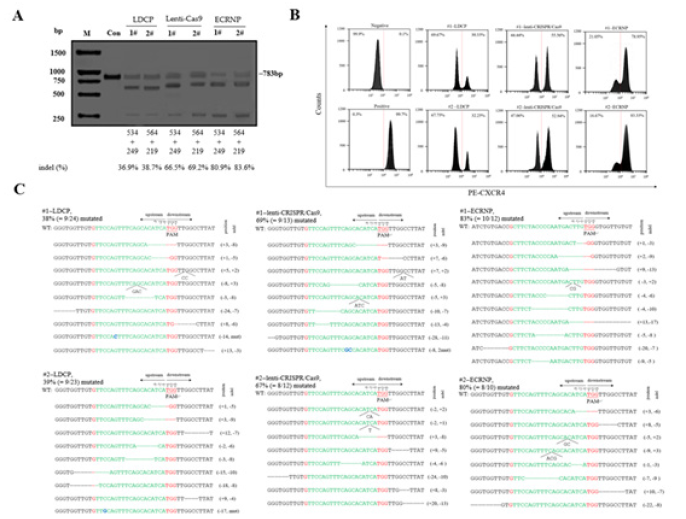
Figure 3: Disruptions of CXCR4 in human CD4+ T cells using three CRISPR/Cas9 systems. (A) Human CD4+ T cells were transfected with CRISPR/Cas9 using three different vectors. After 36 hours, cells were collected for genomic DNA extraction and T7EN1 assay, a method measuring the cleavage efficacy in CXCR4 cut by the three CRISPR/Cas9 systems transported with different vectors. The expected DNA length of the wild-type allele was 783 bp, while the lower bands in lane #1 and #2 indicated mutant CXCR4 alleles. Percentage of indels was determined using Image J software. (B, C) The expression of CXCR4 in the human CD4+ T cell line was analyzed with flow cytometry analysis, followed by calculation of mutation frequencies and on-target analysis of the cleavage on target sites.
Subsequently, flow cytometry analysis was conducted to evaluate the CXCR4 expression in CD4+ T cells using a Cy3-conjugated CXCR4 antibody. These results indicated variations in CXCR4 expression levels in human CD4+ T cells due to CRISPR/Cas9 disruptions delivered via different vectors, potentially influencing protection against HIV-1 infection. Cells collected via CXCR4-based fluorescence-activated cell sorting (FACS) were subjected to genomic DNA extraction. This data suggested that ECRNP efficiently silenced the CXCR4 gene, inducing indels with frequencies of up to 82%, compared to 38% for LDCP and 68% for Lenti-CRISPR/Cas9 (Fig. 3B, C).
Fig. 3. Disruptions of CXCR4 in human CD4+ T cells using three CRISPR/Cas9 systems. (A) Human CD4+ T cells were transfected with CRISPR/Cas9 using three different vectors. After 36 hours, cells were collected for genomic DNA extraction and T7EN1 assay, a method measuring the cleavage efficacy in CXCR4 cut by the three CRISPR/Cas9 systems transported with different vectors. The expected DNA length of the wild-type allele was 783 bp, while the lower bands in lane #1 and #2 indicated mutant CXCR4 alleles. Percentage of indels was determined using Image J software. (B, C) The expression of CXCR4 in the human CD4+ T cell line was analyzed with flow cytometry analysis, followed by calculation of mutation frequencies and on-target analysis of the cleavage on target sites.
CXCR4‑Modified CD4+ T Cells Exhibit Undetectable Off‑Target Effects
It is evident that evaluating potential off-target effects is crucial when considering a gene editing strategy, especially for addressing HIV-1 infection. While CRISPR/Cas9 is a powerful tool for genome manipulation, its precision is paramount, and careful assessment of off-target cleavages is essential to ensure the safety and efficacy of the gene editing approach in diverse research applications. To assess the specificity of CRISPR/Cas9 system in disrupting the CXCR4 gene, we analyzed predicted sgRNA off-target sites (Table 1). Research findings for predicted off-target sites of CXCR4 (sgRNA #1, sgRNA #2) revealed no detectable off-target effects through DNA sequencing for each fragment. This conclusion was further supported by deep sequencing or T7E1 assays conducted on all predicted off-target sites in this study.
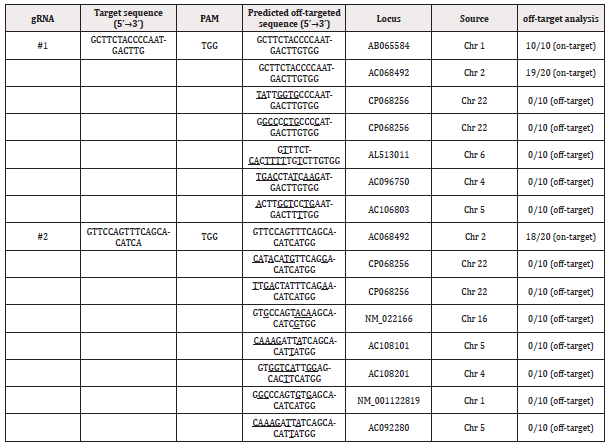
Table 1: Comparative analysis of off-target cleavages in the CXCR4 gene.
Note*: Red letters indicate different bases compared with the on-targeted sequence.
For a comprehensive understanding of potential off-target cleavages, mutation frequency analysis was applied to predicted off-target sites of the CXCR4 gene targeted by the CRISPR/Cas9 system. These off-target sites were predicted and aligned with the human genome published in the NCBI database. Results demonstrated no significant indels at the twelve predicted off-target sites in the genome of all edited cells (Table 1). Gene loci were subsequently amplified, cloned into T-vector, and subjected to sequencing.
Discussion
Antiretroviral therapy, including HAART (highly active antiretroviral drugs), has significantly extended the lifespan of people living with HIV-1 by transforming the AIDS from a life-threatening condition into a chronic disease controlled by lifelong treatment [29]. Indeed, HIV-1 can rapidly and easily develop resistance to antiretroviral drugs [30]. Notably, cases of cures for AIDS, such as the Berlin patient’s case, have served as significant inspiration for researchers. This inspiration has driven the development of genetic engineering tools aiming to confer human cells with resistance to HIV, offering potential alternatives to traditional antiretroviral therapies. Recently, the CRISPR/Cas9 machinery has been developed as a powerful tool for precisely manipulating the human genome, aiming to correct target genes and advance disease treatment. Several studies have explored the use of the CRISPR/Cas9 system to disrupt viral sequences, such as HIV LTR, Gag, Tat, Rev, and pol [31-33]. However, the effectiveness of this method is often hindered by the genomic diversity of HIV-1, resulting in less outcomes [34,35]. In fact, the diversity of HIV-1 is classified into four major groups (M, N, O, and P), exhibiting differences of 25-40% and varying up to 15% marked within-group subtypes [35]. Considering these challenges, researchers have explored that targeting and mutations of vital host genes can also establish host immunity against HIV [36]. Several previous studies have showed that CRISPR/Cas9 system has specificity and high efficiency in targeting CXCR4 exon, rendering cells resistant to HIV-1 infection [37-39].
To effectively delivery the RNA-guided ribonuclease systems into cells and exert its mutation efficacies both in the cytoplasm and in the cellular nucleus, delivery models (e.g., plasmids, lentiviruses, and adeno-associated viruses) have been developed for CRISPR/ Cas9 system. To date, delivery by viral, plasmid, mRNA, and protein-based approaches are most commonly used methods [40]. In the clinical scenario, liposome-mediated gene transfer has suggested promises in targeted gene therapy. An ideal liposomal vector for targeted gene delivery has low toxicity, high stability, small size (<100 nm), neutral surface charge, extended serum half-life (>2-4 h), and moderate immunogenicity [41]. Lipofectamine reagents are widely accepted as “gold-standard” for the safe delivery of exogenous DNA or RNA into cells. However, a comprehensive physiology-based explanation mechanism for their superior efficacy is still absent [42].
In the field of viral-based vectors, three vital strategies are adenoviruses, adeno-associated viruses, and lentiviruses. Despite the potential for undesired random integration into the host genome in most viral systems, lentivirus vectors remain a robust platform for genetically modified cell therapies, which have been successful applied in many clinical trials. These trials have targeted various diseases, including metabolic disorders, immune disorders, cancers, and “orphan” diseases [43]. In essence, effective delivery methods play a crucial role in enabling genome editing systems to precisely target specific genes. Lentiviral vectors, originating from HIV-1, stand out as vital biological delivery systems and have been proven useful for integrating gene information into multiple cell lines [44]. Moreover, these vectors have been engineered to transport therapeutic nucleases, expressing them transiently in gene therapy applications.
Additionally, the application of electroporation technology markedly enhances non-viral gene transfer, either in vitro or in vivo. Electroporation stands out as the primary method for delivering Cas9 ribonucleoprotein (RNP) complexed with guide RNA (gRNA) into the intricate intracellular environment [45].
This approach is considered relatively safe to introduce genes to a wide variety of cell types and tissues, including skeletal muscle, tumor cells, skin cell, nephrocyte and hepatocytes, as evidenced in numerous disease models [46]. The utilization of electroporation for boosting gene delivery is a powerful and useful methodology applicable to both in vitro and in vivo studies because it can induce functional destabilization and permeabilization of cell membranes within the electric field, leading to widespread gene transfer to many cell types [47].
Despite ongoing advancements in delivery systems, achieving targeted and efficacious delivery of complexes of the Cas9 protein and chimeric RNAs remains a critical challenge in applying CRISPR/Cas systems for gene therapy.
While plasmids encoding CRISPR/Cas9 can be efficiently transfected into various cell types, they typically result in only 1-5% ablation of target protein expression in human CD4+ T cells [22]. According to our unpublished experimental data, lentiviral co-delivery of SpCas9 mRNA and expression cassettes encoding a CXCR4-targeted gRNA proves efficacious, inducing over 40% ablation of gene expression. Furthermore, Cas9 ribonucleoprotein (RNP) electroporation has the capacity to enable up to approximately 60% of cells to silence the cell-surface expression of CXCR4, encompassing both knock-out and knock-in mechanisms.
Here in this study, we employed the CRISPR/Cas9 systems packaged in three different transported formats to target and excise CXCR4 gene, aiming to unleash resistant activity against HIV-1 in the T cells. The CRISPR/Cas9-mediated deletion of CXCR4 potentially offers an alternative way to treat the disease of X4-tropic HIV- 1 infection. Advances in vector construction technology accelerates precise release and functional development of CRISPR/Cas9 apparatus. Generally, optimal exogenous vector of gene delivery exhibits high payload capacity and cell tropism, high transduction efficiency, little or no genotoxicity and cytotoxicity, and triggers minimal immunity response [48]. Several successful clinical tests have already been reported by applying adenoviral vectors for genome editing of CCR5 in AIDS, herpesvirus vectors for cancer, and adeno-associated virus vectors for hemophilia [49]. Simultaneously, organ-specific targeting of CRISPR/Cas9 delivery vehicles is a challenge that has recently gained prominence in gene editing research. To some certain, for targeted delivery of Cas9 nucleases and gRNA to therapeutically relevant cell types in gene therapy, the correct vector selection may be second only to the design and screen of gRNA.
In our investigations, the sequencing data showed that some edited CXCR4 genes via CRISPR/Cas9 system did not result in the loss of CXCR4 expression. Therefore, these mutant CXCR4 genes can be assigned to the “invalid editing”. Additionally, off-target analysis indicated that no evident off-target mutagenesis has been observed in any of the predicted sites in primary CD4+ T cells, confirming the specificity of two deployed sgRNAs.
Notably, studies have demonstrated that embryos with CXCR4 deficiencies struggle to develop and survive, with accidental survivors inevitably develop warts, hypogammaglobulinemia, infections, and myelokathexis syndrome [50]. However, in our study, the edited cells with CXCR4 gene modification did not show obvious obstacles to growth and development. These findings support the observation that CRISPR/Cas9 editing system is a powerful technique to knockout HIV restriction gene of host cell in vitro with low off-targeting efficiency.
AIDS remains an incurable disease, although the occurrence of cases of cure, treated with special treatment or without any additional therapies, have been reported globally. Despite this, these cured cases provide hope for developing strategies to achieve HIV sterilization in human body. Gene therapy strategies hold great promising in the treatment of HIV and attaining its functional treatment. Studies on CRISPR/Cas-edited HIV co-reporter genes are important for understanding the detailed mechanism and developing countermeasures for the prevention and control of AIDS/HIV infection. Additionally, CXCR4-related studies are important for understanding the underlying mechanisms and developing measures for the prevention and control of other human diseases. Drug and gene intervention studies on CXCR4 have special value for elucidating the internal relationships between genotype and phenotype.
However, a major challenge to gene therapy-based treatment of HIV lies in finding suitable therapeutic modes (DNA, mRNA, or RNP) and reliable vehicles (viral or non-viral) to induce new host immunity for fighting against infections of heterogenous HIV strains.
Conclusions
As an endogenous HIV restriction factor, the immune balance within the human body may contribute to eradicate HIV infection. To date, the emergence of CRISPR/Cas9-based genome editing technology has renewed investigation into appropriate delivery vehicles. In summary, our study is a proof-of-concept, demonstrating a holistic immunity against HIV-1 infection via CRISPR/Cas9 mediated host’s specific gene editing of CXCR4, even though its molecular mechanism is still not clear. Together, these findings highlight the importance of developing appropriate vectors is a synergistic agent to increase therapeutic efficiency for human CD4+ T cells to block HIV-1 infection in CRISPR/Cas9-based treatment of HIV/ AIDS. Future studies focusing on relevant vectors are essential to identify the optimal delivery strategy of CRISPR therapeutics.
Author Contributions
Conceptualization, H.F.L., G.L. and L.S.; methodology, H.F.L. and X.W.P.; software, G.L.; validation, H.F.L. and H.Z.W.; formal analysis, G.L. and H.Z.W.; writing-original draft preparation, H.F.L. and G.L.; writing-review and editing, X.W.P. and H.F.L.; supervision, L.S. and L.Y.; project administration, H.F.L. and M.J.R.; funding acquisition, X.W.P., and L.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research supported by Clinical Medical Technology Demonstration Base for Genetic Research of Fetal Congenital Heart Disease in Hunan Province (2021SK4036) , Hunan Province Children’s Safe Medication Clinical Medical Technology Demonstration Base(2023SK4083), Project of Changsha Science and Technology Bureau, (KH2201045), Natural Science Foundation of Hunan Province (2023JJ30063), Changsha Science and Technology Bureau natural science surface project (kq2202030) and National Natural Science Foundation of China(32070817).
Acknowledgments
We wish to thank PhD Qiudi Deng and college teacher Xun Chen for providing constructive suggestions for this research.
Conflicts of Interest
The authors declare to have no conflicts of interests regarding the publication of this paper.
References
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, et al. (2013) RNA-guided human genome engineering via Cas9. Science 339(6121): 823-826.
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, et al. (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8(11): 2281-2308.
- Nazari R, Joshi S (2008) CCR5 as target for HIV-1 gene therapy. Curr Gene Ther 8(4): 264-272.
- Ray N, Doms RW (2006) HIV-1 co-receptors and their inhibitors. Current Topics in Microbiology and Immunology. Curr Top Microbiol Immunol 303: 97-120.
- Wilen CB, Wang J, Tilton JC, Miller JC, Kim KA, et al. (2011) Engineering HIV-resistant human CD4+ T cells with CXCR4-specific zinc-finger nucleases. PloS Pathogens 7(4): e1002020.
- Hütter G, Bodor J, Ledger S, Boyd M, Millington M, et al. (2015) CCR5 Targeted Cell Therapy for HIV and Prevention of Viral Escape. Viruses 7(8): 4186-4203.
- Marmor M, Sheppard HW, Donnell D, Bozeman S, Celum C, et al. (2001) Homozygous and heterozygous CCR5-Delta32 genotypes are associated with resistance to HIV infection. Journal of Acquired Immune Deficiency Syndrome 27(5): 472-481.
- Murdoch C (2000) CXCR4: chemokine receptor extraordinaire. Immunological Reviews 177: 175-184.
- Saayman S, Ali SA, Morris KV, Weinberg MS (2015) The therapeutic application of CRISPR/Cas9 technologies for HIV. Expert Opinion on Biological Therapy. Expert Opin Biol Ther 15(6): 819-830.
- Wang Z, Pan Q, Gendron P, Zhu W, Guo F, Cen S, et al. (2016) CRISPR/Cas9-Derived Mutations Both Inhibit HIV-1 Replication and Accelerate. Viral Escape. Cell Reports 15(3): 481-489.
- Wang G, Zhao N, Berkhout B, Das AT (2016) CRISPR-Cas9 Can Inhibit HIV-1 Replication but NHEJ Repair Fa-cilitates Virus Escape. Molecular Therapy. Mol Ther 24(3): 522-526.
- Wang Z, Wang W, Cui YC, Pan Q, Zhu W, et al. (2018) HIV-1 Employs Multiple Mechanisms to Resist Cas9/Single Guide RNA Targeting the Viral Primer Binding Site. Journal of Virology. J Virol 92(20): e01135-18.
- Darcis G, Binda CS, Klaver B, Herrera-Carrillo E, et al. (2019) The Impact of HIV-1 Genetic Diversity on CRISPR-Cas9 Antiviral Activity and Viral Escape. Viruses 11(3): 255.
- Yoder KE, Bundschuh R (2016) Host Double Strand Break Repair Generates HIV-1 Strains Resistant to CRISPR/Cas9. Scientific Reports 6: 1-12.
- Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, et al. (2014) Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers re-sistance to HIV infection. Proceedings of the National Academy of Sciences of the United States of America. Proc Natl Acad Sci U S A 111(26): 9591-9596.
- Li C, Guan X, Du T, Jin W, Wu B, Liu Y, et al. (2015) Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. Journal of General Virology. J Gen Virol 96(8): 2381-2393.
- Xu L, Yang H, Gao Y, Chen Z, Xie L, Liu Y, et al. (2017) CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance In Vivo. Molecular Therapy. Mol Ther 25(8): 1782-1789.
- Hou P, Chen S, Wang S, Yu X, Chen Y, Jiang M, et al. (2015) Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Scientific Reports. Sci Rep 5: 15577.
- Lebbink RJ, de Jong DC, Wolters F, Kruse EM, van Ham PM, et al. (2017) A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Scientific Reports. Sci Rep 7: 41968.
- Turk G, Seiger K, Lian X, Sun W, Parsons EM, Gao C, et al. (2022) A Possible Sterilizing Cure of HIV-1 Infection Without Stem Cell Transplantation. Annals of Internal Medicine. Ann Intern Med 175(1): 95-100.
- Hultquist JF, Schumann K, Woo JM, Manganaro L, McGregor MJ, et al. (2016) A Cas9 Ribonucleoprotein Platform for Functional Genetic Studies of HIV-Host Interactions in Primary Human T Cells. Cell Reports 17(5): 1438-1452.
- Mandal PK, Ferreira LM, Collins R, Meissner TB, Boutwell CL, Friesen M, et al. (2014) Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 15(5): 643-652.
- Xiao Q, Chen S, Wang Q, Liu Z, Liu S, et al. (2019) CCR5 editing by Staphylococcus aureus Cas9 in human primary CD4+ T cells and hematopoietic stem/progenitor cells promotes HIV-1 resistance and CD4+ T cell enrichment in humanized mice. Retrovirology 16(1): 15.
- Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, et al. (2015) Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proceedings of the National Academy of Sciences of the United States of America. Proc Natl Acid Sci U S A 112(33): 10437-10442.
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343(6166): 84-87.
- Liu Z, Chen S, Jin X, Wang Q, Yang K, Li C, et al. (2017) Genome editing of the HIV co-receptors CCR5 and CXCR4 by CRISPR-Cas9 protects CD4+ T cells from HIV-1 infection. Cell Bioscience 7: 47.
- Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, et al. (2000) Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by co-receptor specificity defined by the V3 loop of gp120. Journal of Virology. J Viral 74(18): 8358-8367.
- Klase Z, Yedavalli VS, Houzet L, Perkins M, Maldarelli F, et al. (2014) Activation of HIV-1 from latent infection via synergy of RUNX1 inhibitor Ro5-3335 and SAHA. PLoS Pathog 10(3): e1003997.
- Yeni P (2006) Update on HAART in HIV. Journal Of Hepatology. J Hepatol 44(1 Suppl): S100-S103.
- Clutter DS, Jordan MR, Bertagnolio S, Shafer RW (2016) HIV-1 drug resistance and resistance testing. Infection Genetics and Evolution. Infect Genet Evol 46: 292-307.
- Ueda S, Ebina H, Kanemura Y, Misawa N, Koyanagi Y (2016) Anti-HIV-1 potency of the CRISPR/Cas9 system insufficient to fully inhibit viral replication. Microbiology and Immunology. Microbiol Immunol 60(7): 483-496.
- Yin L, Hu S, Mei S, Sun H, Xu F, Li J, et al. (2018) CRISPR/Cas9 inhibits multiple steps of HIV-1 infection. Human Gene Therapy. Hum Gene Ther 29(11): 1264-1276.
- Herskovitz J, Hasan M, Patel M, Blomberg WR, Cohen JD, et al. (2021) CRISPR-Cas9 Mediated Exonic Disruption for HIV-1 Elimination. EBioMedicine 73: 103678.
- Bunnell BA (2021) Excision of latent HIV-1: CRISPR technology overcomes viral strain diversity. EBioMedicine 74: 103720.
- Roychoudhury P, De Silva Feelixge H, Reeves D, Mayer BT, Stone D, et al. (2018) Viral diversity is an obligate consideration in CRISPR/Cas9 designs for targeting the HIV reservoir. BMC Biology 16(1): 75.
- Sessions KJ, Chen YY, Hodge CA, Hudson TR, Eszterhas SK, Hayden MS, et al. (2020) Analysis of CRISPR/Cas9 Guide RNA Efficiency and Specificity Against Genetically Diverse HIV-1 Isolates. AIDS Research and Human Retroviruses 36(10): 862-874.
- Wang Q, Chen S, Xiao Q, Liu Z, Liu S, Hou P, et al. (2017) Genome modification of CXCR4 by Staphylococcus aureus Cas9 renders cells resistance to HIV-1 infection. Retrovirology 14(1): 51.
- Liu S, Wang Q, Yu X, Li Y, Guo Y, Liu Z, et al. (2018) HIV-1 inhibition in cells with CXCR4 mutant genome created by CRISPR-Cas9 and piggyBac recombinant technologies. Scientific Reports. Sci Rep 8(1): 8573.
- Luther DC, Lee YW, Nagaraj H, Scaletti F, Rotello VM (2018) Delivery approaches for CRISPR/Cas9 therapeutics in vivo: advances and challenges. Expert opinion on drug delivery 15(9): 905-913.
- Knipping F, Newby GA, Eide CR, McElroy AN, Nielsen SC, Smith K, et al. (2022) Disruption of HIV-1 co-receptors CCR5 and CXCR4 in primary human T cells and hematopoietic stem and progenitor cells using base editing. Molecular therapy: the journal of the American Society of Gene Therapy. Mol Ther 30(1): 130-144.
- Zylberberg C, Gaskill K, Pasley S, Matosevic S, et al. (2017) Engineering liposomal nanoparticles for targeted gene therapy. Gene Therapy 24(8): 441-452.
- Cardarelli F, Digiacomo L, Marchini C, Amici A, Salomone F, Fiume G, et al. (2016) The intracellular trafficking mechanism of Lipofectamine-based transfection reagents and its implication for gene delivery. Scientific reports. Sci Rep 6: 25879.
- Bulcha JT, Wang Y, Ma H, Tai P, Gao G (2021) Viral vector platforms within the gene therapy landscape. Signal transduction and targeted therapy. Signal Transduct Target Ther 6(1): 53.
- Schambach A, Zychlinski D, Ehrnstroem B, Baum C (2013) Biosafety features of lentiviral vectors. Human gene therapy. Hum Gene Ther 24(2): 132-142.
- Hamilton JR, Tsuchida CA, Nguyen DN, Shy BR, McGarrigle ER, Sandoval Espinoza CR, et al. (2021) Targeted delivery of CRISPR-Cas9 and transgenes enables complex immune cell engineering. Cell Reports 35(9): 109207.
- Gehl J (2003) Electroporation: theory and methods, perspectives for drug delivery, gene therapy and research. Acta physiologica Scandinavica 177(4): 437-447.
- Dean DA (2013) Cell-specific targeting strategies for electroporation-mediated gene delivery in cells and animals. The Journal of membrane biology. J Membr Biol 246(10): 737-744.
- Nayerossadat N, Maedeh T, Ali PA (2012) Viral and nonviral delivery systems for gene delivery. Advanced biomedical research. Adv Biomed Res 1: 27.
- Athanasopoulos T, Munye MM, Yáñez-Muñoz RJ (2017) Nonintegrating Gene Therapy Vectors. Hematology/oncology clinics of North America. Hematol Oncol Clin North Am 31(5): 753-770.
- Peng Y, Yang T, Tang X, Chen F, Wang S (2020) Construction of an Inducible CRISPR/Cas9 System for CXCR4 Gene and Demonstration of its Effects on MKN-45 Cells. Cell Biochemistry and Biophysics. Cell Beachem Biophys 78(1): 23-30.



 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.