Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Comparison of AI-Based and Manual Reader Evaluation of Biparametric MRI of Prostate with Raised PSA Levels
*Corresponding author: Atul Kapoor, Consultant Radiologists Advanced Diagnostics and Institute of Imaging, Amritsar, India.
Received: November 02, 2024; Published: November 11, 2024
DOI: 10.34297/AJBSR.2024.24.003237
Abstract
Objectives: Cancer prostate is the most common cancer in males worldwide and early diagnosis has been a challenge since estimation of blood Prostate Surface Antigen (PSA) levels is non-specific to establish the diagnosis and yet continues to be the mainstay test. Of late Biparametric MRI (bpMRI) has been suggested as the second line test before doing a biopsy to determine the likelihood of a prostatic cancer. This study was designed to determine if use of Artificial intelligence (AI) can improve the results of bpMRI in diagnosis and plan targeted biopsies. The study compared the results of the use of AI with manually read studies by radiologists.
Methods: 50 patients with raised PSA level of more than 4ng/ml underwent bp MRI. The studies were independently evaluated by two experienced radiologists with one formally trained for prostatic evaluation. AI based prostate evaluation was done later and the results compared for diagnostic performance and lesion localization and PIRAD v2 categorization. Targeted biopsy under ultrasound and shear wave elastography was done using trans rectal approach and final diagnosis made giving Gleason score and Gleason grade group.
Results: AI showed highest accuracy to detect both Pca and Cs Pca nodules of 91.1% and 100% on per patient basis respectively with improved negative predictive value of 93% compared to manually read studies. AI detected iPca nodules with sensitivity and specificity of 91% each.
Conclusion: Use of AI improves the diagnostic performance of bp MRI than manually read examinations and shows high negative predictive value which is of use to prevent unnecessary biopsies.
Keywords: Biparametric MRI, Prostate cancer, Clinically significant cancer, Elastography, Targeted biopsy
Introduction
Cancer prostate is the most prevalent cancer in men worldwide [1]. Cancer screening for prostate cancer has been commonplace with the use of PSA blood levels and a cut-off of 4ng/ml has been suggested as the threshold for biopsy [2]. However, 27-54% of patients with high PSA have been shown to have either no cancers or clinically insignificant nodules while 15-44% of cancer patients may have normal PSA levels [3]. Hence there was a need for an accurate imaging tool and biparametric - bpMRI has been suggested as the examination of choice. PROMIS and PRECISION trials have shown that the use of bpMRI can not only reduce the number of unnecessary biopsies but can help plan targeted biopsies of the prostate increase accuracy and reduce complications [4,5]. Artificial Intelligence (AI) is being increasingly used in imaging workflows nowadays to optimize and reduce turnaround evaluation times [6,7]. This study was planned to evaluate the use of AI-based evaluation of bpMRI data of patients with raised PSA levels who underwent biopsy studies and to compare results with manually read radiologist findings.
Material and Methods
The study comprised of 50 consecutive patients with raised PSA levels of more than 4ng/ml who came to us for further evaluation and ultrasound guided biopsy. After informed consent and approval of the study from the local ethics committee all patients underwent bpMRI examination using a standard protocol.
The primary purpose of this study was to compare the diagnostic performance of AI-based evaluation of bpMRI of the prostate with two reader evaluation groups i.e. R1–routine experienced radiologist and R2 multiparametric trained radiologist- R2 for detection of prostate carcinoma in patients with raised PSA. The Inclusion criteria included patient’s biopsy naïve patients with raised PSA. Exclusion criteria were patients with acute urinary infection, prior known patients of carcinoma prostate, or patients with contradiction of MRI procedure. The final diagnosis was confirmed based on biopsy results with estimation of the Gleason score and Gleason grade group for each patient.
MRI Examination
MRI examination was done on 1.5 tesla Siemens Amira system using an abdominal flex coil with the patient in the supine position. Standard T2W and T1WI axial and coronal sequences for the prostate were used as enlisted in the DOT workflow for the prostate in the system with a small Field of view followed by high-resolution Resolve diffusion sequences in the axial plane using high b value of at least 1400 along with ADC reconstructed maps. The images were transferred to the Syngo-via workstation and evaluated by experienced radiologist for any nodule in the Transitional, peripheral, central zones on both T2WI and DWI using the PIRAD v2.0 lexicon.
The second radiologist then used an AI-based deep learning commercial algorithm Prostate MRI (Siemens AG) to detect the nodules all which were colour coded along with size of the index nodule by the software and each graded using PIRAD V2 grading system. All nodules were recorded for size i.e. volume, ADC value on DWI along with overall volume of prostate and PSA density also estimated.
TRUS-Guided Biopsy
Transrectal ultrasound was done after reviewing the MRI images. The examination was done in left lateral decubitus position on Aplio system (Canon USA) after observing necessary antibiotic prophylaxis. Standard endocavitary probe covered with sheath and biopsy guide was introduced into the rectum and axial and sagittal images of prostate obtained on grey scale. The shear wave elastogram was then done using 2D shear wave technology and the area of focal stiffness visualized and correlated with the nodule seen on the bpMRI. 18 G 20 cm automatic trucut biopsy needle (Bard USA) was introduced and four 4 cm length biopsies obtained from the site of stiffness. Two biopsies were taken from contralateral lobe for baseline characteristics. All biopsies were reported in standard Gleason score format with length of positive biopy cores and findings summarized into Gleason grade group. Patients with GGG2 or above or with Gleason score 3+4 and above labelled as having clinically significant PCa (csPCA) while those with Gleason score of 3+3 or less / GGG1 were labelled as clinically insignificant/indolent I PCa nodules (Figure 1).
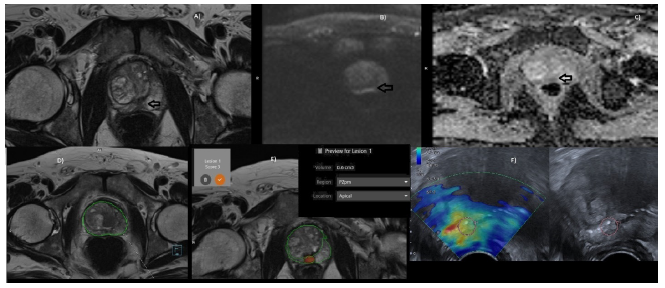
Figure 1: (A-F): A) Plain T2W axial image of prostate showing focal hypointense lesion in right Median PZ (arrow). B) Diffusion image showing hyperintense lesion at the same location(arrow). C) ADC showing restricted ADC of 1.04 (arrow). D) AI segmentation of prostate with volume of 51cm3 E) AI- labeled PIRAD 3 nodule of 0.6cm3. F) Elastographic TRUS guided biopsy with ROI showing target lesion.
MRI Image Reading
It was done by R1 and R2 radiologists on Siemens workstation (syngo via) independently and the reporting was structured as per PI-RADS v2.0. Both were blinded to the results of each other and of the findings on AI program. Later the same data was evaluated using Siemens AI- Prostate in which prostate segmentation, volume estimation, and nodule detection were done with categorization of prostatic nodule as per PIRADS v2.0 along with detection of index lesion. All patients with a PIRADS score of 3.0 or more were deemed as positive for Pca
Statistical Analysis
Was done using Analyse-IT software. All continuous variables were evaluated for mean values with standard deviation. Sensitivity, specificity, positive and negative predictive values, and AUCs were calculated based on the ground truth of biopsy results for both AI-based and manually read findings based on PIRADS categories. PIRADS 3 and > lesions were labelled positive for presence of PCa on imaging. Gleason 3+4 above. Gleason grade group II and above was labeled as being positive for clinically significant cancer (csPca). Patients with Gleason score 3+3=6 i.e. Gleason grade group I was categorized as having indolent/clinically insignificant cancer (iPCa). Accordingly, a case was labelled as negative when only PI-RADS 1 and 2 scores were reported. Per-lesion and index level, sensitivities were calculated kappa statistics were applied to compare the agreement concerning the PI-RADS. The k coefficients were assessed as follows: 0.01–020, slight agreement; 0.21–0.40, fair agreement; 0.41–0.60, moderate agreement; 0.61–0.80, substantial agreement; and 0.81–0.99, almost perfect agreement. p values <0.05 were considered significant.
Results
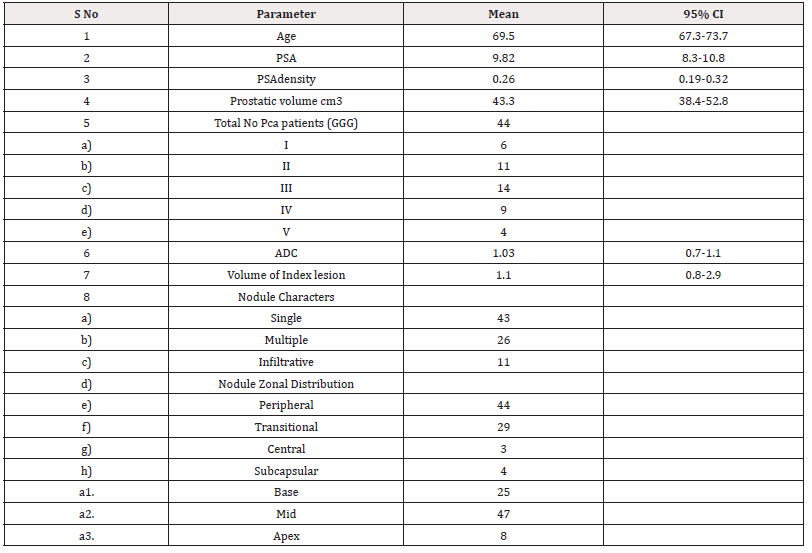
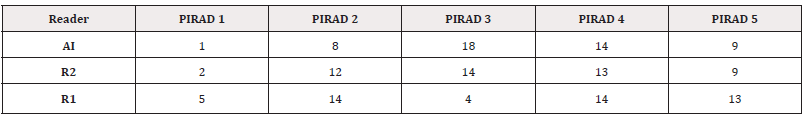
This study comprised 50 patients who presented to us with raised PSA, mean 9.82ng/ml (95% CI 8.7 – 10.85). The mean age was 69.5 years (95% CI 67.3–71.7). The mean PSA density was 0.26 (95% CI 0.19 – 0.32). The other demographics are given in Table 1. A total of 42 patients had prostatic cancer nodules on biopsy examination the Gleason grade grouping is given in Table 1. There were 17 patients with indolent prostatic cancer while 27 patients had clinically significant cancer nodules. A total of 80 nodules were present, AI detected 76 while Readers 1 and 2 detected 43 and 64 nodules which were assigned PIRAD v2.0 score with PIRAD 3 being the largest detected by AI and R2 (Table2).
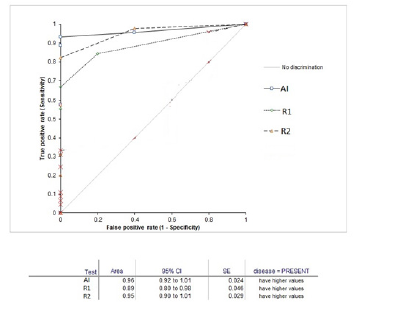
The mean volume of the index lesion was 1.1 cm3. The pattern of nodules was multiple in 43 nodules (Figure 2), 26 were single and 11 were infiltrative (Figure 3). The most common pattern of nodule distribution was seen in the peripheral zone [4] while subcapsular and central zone nodules were the least common [4]. The apical part of the prostate showed the least common presence of nodules [8]. AI detected a total of 38 true positive patients of Pca with three false positives while 1 patient was missed by AI and was seen in a subcapsular location (Figure 4). The 3 false positives were all in the PIRAD 3 group. Reader 2 detected 36 patients with prostatic cancer while only 31 patients were detected by reader with Pca. On a per-patient basis AI, Reader 1, and Reader 2 diagnosed patients with cancer nodules with sensitivities of 95.4 %, 87.5%, 79.20% and specificities of 72.7%, 80.7% and 66.6% with an accuracy of 91.1%,86% a nd 76.9% respectively (Table 3). A total of 80 nodules were detected and on per nodule basis the sensitivity, specificity, and accuracy were 94%, 100%, 95% for AI while it was 81%, 65%, 77% for R2 and,62 %, 63% and 62% for R1 (Table 4). A moderate inter-observer agreement was seen between both readers with a Kappa coefficient of 0.58 while a weak agreement was seen between AI and both readers (kappa =0.37). 31 patients had csPca. The AUC's analysis done for AI, R2 and R1 was0.96, 0.95 and 0.89 respectively (Figure 5). There were 31 patients with clinically significant prostate cancer on biopsy and 11 patients with iPca (Figure 6). AI had 100% diagnostic performance to detect csPca while R2 and R1 showed sensitivity and specificity was 86%, 81%, and 81%, and 37% with NPV of 68% and 62% respectively (Table 5). AI had 91.1% sensitivity and specificity to detect iPca lesions. Out of the 11 iPca patients, 8 patients were PIRAD 3 while 3 were in PIRAD 4 category.

Figure 2: (A-H): A) T2W axial image showing hypodense nodules in left PZm in mid part B-C) DWI and ADC images showing increased signal of nodule with restricted ADC 9.6. D) AI- analysis with labeled Pca nodules in PZm in left lobe with max volume of 1.8cm3- PIRAD 5. E) Basal part of gland showing hypointense nodule in TZa. F-G) DWI and ADC images of the same lesion.
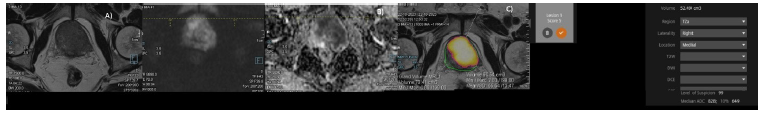
Figure 3: (A-D): A) T2W axial image in GG grade 4 group with CsPCA showing diffuse hypointense signal of prostate with loss of zonal anatomy. B-C) DWI and ADC images showing diffuse hyperintense signal with restricted ADC of 0.86 D) AI showing infiltrative nodule with volume of 52 cm3 of index lesion – PIRAD 5.
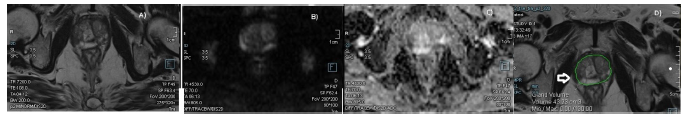
Figure 4: (A-D): A) T2W axial image showing hypointense subcapsular lesions in both lobes in apical PZPL zones. B-C) DWI and ADC images showing hyperintense signal with restricted ADC with mean of 1.1 –PIRAD 3; GG 2 group IPca. D) AI- Image showing segmentation of prostate with volume of 51cm3 with segmentation excluding the areas of interest with no lesion detected (arrow).
Discussion
Our study showed that 88% of men with raised PSA more than 8nmg/ml and raised PSA density above 0.20 had Pca. Out of these 70.5% of patients with raised PSA (between 8-10ng/ml) had csPCA while 29.5% of patients had PCA. Similar incidence has been reported by prostate cancer prevention trial [8]. In the present study bpMRI studies read by AI improved the accuracy to 91% from 62% and 74% of both the radiologist groups -R1,2 with improved AUCs. Similar results were published by Winkel, et al. [9] who showed that of the AI model improved the radiologist's performance for locating MRI-visible lesions in terms of an increase in AUC from 0.84 to 0.88. Winkel, et al. [9] did report an improved inter-observer analysis with AI use however present study showed only a fair kappa analysis of 0.38. These were in agreement with results by Cipollari, et al. [7] of Inter-reader agreement concerning mpMRI was moderate for bpMRI, Quantib, and qADC for both the inexperienced (weighted k of 0.42, 0.45, and 0.41, respectively) and the experienced radiologists (weighted k of 0.44, 0.46, and 0.42, respectively). Cuocolo, et al. [10] did a metanalysis of 12 studies to detect csPCA using AI with ML or deep learning AI models and also showed an AUC's of 0.86 and suggested its use for bpMRI. Suarez-Ibarrola, et al. [11] also suggested after a review of 11 studies that validated AI bpMRI studies had a high sensitivity of 92% to detect csPCA patients and recommended its use. Similar results were shown by the current study in detecting csPCA patients with an accuracy of 100% for this group. The current study showed an incidence of 25% cases of iPCA the diagnosis of which forms a formidable challenge. Most i.e. 8/11 patients were in the PIRAD 3 category which forms the indeterminate nodule group similar to results reported in the literature [12-13].
AI group showed a high negative predictive value of 93% per patient which suggested that this was appropriate to be used to determine the need for a targeted biopsy in patients with high PSA. In patients with a normal bpMRI, a biopsy can be postponed. Detection of iPca is important not only from a diagnosis point of view but also from a treatment viewpoint as these patients are unnecessarily subjected to radical overtreatment instead of active surveillance. Similar observations were made in a systemic review by Penzkofer, et al. [14] and in a joint position paper by ESUR where it was shown that the use of AI to triage prostatic nodules is an accurate tool and it can accurately differentiate between CSPca from IsPCA nodules. Similar results were also shown in the PI-PCAi study by Saha, et al. [15] and Sushentsev, et al. [16] where AI achieved an AUC of 0.92 which was higher than the results of pooled data from 62 radiologists. In the present study, only one case was not detected by AI where the patient had bilateral subcapsular PCA nodules in the apical region which was labeled by R2 as a PIRAD 3 lesion and on biopsy was Gleason score 3+3 in GG I group. The most likely reason for false negatives in these cases could be the segmentation error of the prostate gland by AI as is shown in the case example.
Westephalen, et al. [17] also showed in their study that AI missed lesions on T2 images in the apical region of the prostate and showed an accuracy of 89% but AI obviated the high variability in the positive predictive value of 35-49% by routine radiologists with weak inter-observer correlation amongst radiologists which was similar to results observed by us. 8 patients in PIRAD 3 group labeled by AI had IPca and were in GGG I while 3 patients were had no cancer on targeted biopsy. Since the majority of patients in the PIRAD 3 group of the present study had iPCA the results strengthen our protocol of keeping PIRAD 3 category lesions and agree with this criteria as suggested by the PI-PCAi study where they kept the criteria of PIRAD 3 and above as those with Pca. We agree that even though as per the definition the PIRAD 3 nodules are indeterminate and hence cannot be labeled as those with Pca this would lead to unnecessary biopsies, add patient anxiety, and increase false positive cases with reduced sensitivity. Labeling all cases of PIRAD as no cancer would also increase the risk of missing PCa and not putting them in active surveillance. This is more detrimental, especially in countries where there is no formal prostate screening programme.
Schoot, et al. [18] also suggested that it is prudent to keep PIRAD 3 and above as cut off and in such cases, one should upgrade the lesion by combining with multi-variate risk models or even use dynamic CE studies [19,20].
In conclusion, our study shows that AI shows higher diagnostic performance than manually read studies by routine and bpMRI-trained radiologists in patients with moderately raised PSA and high PSA density biopsy naïve patients to detect both csPCa and iPCa lesions and forms a useful tool to plan targeted biopsies. The potential limitations of the study are a) single-center study with only 50 patients with no follow-up data of the patient’s following diagnosis. b) Targeted biopsies were performed using the shear wave transrectal method rather than fusion biopsies which are the gold standard.
Acknowledgement
None.
Conflict of Interest
None.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, et al. (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3): 209-249.
- Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, et al. (2009) Lead Time and Overdiagnosis in Prostate-Specific Antigen Screening: Importance of Methods and Context. J Natl Cancer Inst 101(6): 374-383.
- Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, et al. (2004) Prevalence of Prostate Cancer among Men with a Prostate-Specific Antigen Level < or = 4.0 Ng per Milliliter. N Engl J Med 350(22): 2239-2246.
- Ahmed HU, El Shater Bosaily A, Brown LC, Gabe R, Kaplan R, et al. (2017) Diagnostic Accuracy of Multi-Parametric MRI and TRUS Biopsy in Prostate Cancer (PROMIS): A Paired Validating Confirmatory Study. Lancet 389(10071): 815-822.
- Kasivisvanathan V, Rannikko AS, Borghi M, Panebianco V, Mynderse LA, et al. (2018) MRI-Targeted or Standard Biopsy for Prostate-Cancer Diagnosis. N Engl J Med 378(19): 1767-1777.
- Giannini V, Mazzetti S, Armando E, Carabalona S, Russo F, et al. (2017) Multiparametric Magnetic Resonance Imaging of the Prostate with Computer-Aided Detection: Experienced Observer Performance Study. Eur Radiol 27(10): 4200-4208.
- Cipollari S, Pecoraro M, Forookhi A, Laschena L, Bicchetti M, et al. (2022) Biparametric prostate MRI: impact of a deep learning-based software and of quantitative ADC values on the inter-reader agreement of experienced and inexperienced readers. Radiol Med 127(11): 1245-1253.
- Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, et al. (2009) Screening and prostate-cancer mortality in a randomized European study. N Engl J Med 360(13): 1320-1328.
- Winkel DJ, Tong A, Lou B, Ali Kamen, Dorin Comaniciu, et al. (2021) A novel deep learning based computer-aided diagnosis system improves the accuracy and efficiency of radiologists in reading biparametric magnetic resonance images of the prostate: results of a multireader, multicase study. Invest Radiol 56(10): 605-613.
- Cuocolo R, Cipullo MB, Stanzione A, Valeria Romeo, Roberta Green, et al. (2020) Machine learning for the identification of clinically significant prostate cancer on MRI: a meta-analysis. Eur Radiol 30(12): 6877-6887.
- Borkowetz A, Platzek I, Toma M, Theresa Renner, Roman Herout, et al. (2017) Evaluation of Prostate Imaging Reporting and Data System Classification in the Prediction of Tumor Aggressiveness in Targeted Magnetic Resonance Imaging/Ultrasound- Fusion Biopsy. Urol Int 99(2): 177-185.
- Borkowetz A, Hadaschik B, Platzek I, Marieta Toma, Georgi Torsev, et al. (2018) Prospective comparison of transperineal magnetic resonance imaging/ ultrasonography fusion biopsy and transrectal systematic biopsy in biopsy-naïve patients. BJU Int 121(1): 53-60.
- Filson CP, Natarajan S, Margolis DJ, Jiaoti Huang, Patricia Lieu, et al. (2016) Prostate cancer detection with magnetic resonance-ultrasound fusion biopsy: The role of systematic and targeted biopsies. Cancer 122(6): 884-892.
- Penzkofer T, Padhani AR, Turkbey B, Haider MA, Huisman H, et al. (2021) ESUR/ESUI position paper: developing artificial intelligence for precision diagnosis of prostate cancer using magnetic resonance imaging. Eur Radiol 31(12): 9567-9578.
- Anindo Saha, Joeran S Bosma, Jasper J Twilt, Bram van Ginneken, Anders Bjartell, et al. (2024) Artificial intelligence and radiologists in prostate cancer detection on MRI (PI-CAI): an international, paired, non-inferiority, confirmatory study. The Lancet Oncol 25(7): 879-887.
- Sushentsev N, Caglic I, Sala E, Nadeem Shaida, Rhys A Slough, et al. (2020) The effect of capped biparametric magnetic resonance imaging slots on weekly prostate cancer imaging workload. Br J Radiol 93(1108): 20190929.
- Westphalen AC, McCulloch CE, Anaokar JM, Sandeep Arora, Nimrod S Barashi, et al. (2020) Variability of the positive predictive value of PI-RADS for prostate MRI across 26 centers: experience of the Society of Abdominal Radiology Prostate Cancer Disease-Focused Panel. Radiology 296(1): 76-84.
- Schoots IG (2018) MRI in Early Prostate Cancer Detection: How to Manage Indeterminate or Equivocal PI-RADS 3 Lesions? Transl Androl Urol 7(1): 70-82.
- Lai WS, Gordetsky JB, Thomas JV, Jeffrey W Nix, Soroush Rais Bahrami (2017) Factors predicting prostate cancer upgrading on magnetic resonance imaging-targeted biopsy in an active surveillance population. Cancer 123(11): 1941-1948.
- Kaufmann S, Bedke J, Gatidis S, J Hennenlotter, U Kramer, et al. (2016) Prostate cancer gene 3 (PCA3) is of additional predictive value in patients with PI-RADS grade III (intermediate) lesions in the MR-guided re-biopsy setting for prostate cancer. World J Urol 34(4): 509-515.







 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.